Chapter 8 Data normalization
Data normalization (feature scaling) is not always needed for e.g. decision-tree-based models.
Data normalization is beneficial for:
- Support Vector Machines, K-narest neighbors, Logistic Regression
- Neural networks
- Clustering algorithms (K-means, K-medoids, DBSCAN, etc.)
- Feature extraction (Principal Component Analysis, Linear Discriminant Analysis, etc)
Min-max scaling
- Maps a numerical value \(x\) to the [0,1] interval
\[x' = \frac{x - min}{max - min}\]
- Ensures that all features will share the exact same scale.
- Does not cope well with outliers.
Standardization (Z-score normalization)
- Maps a numerical value to \(x\) to a new distribution with mean \(\mu = 0\) and standard deviation \(\sigma = 1\)
\[x' = \frac{x - \mu}{\sigma}\]
- More robust to outliers then min-max normalization.
- Normalized data may be on different scales.
Example
library(dplyr)
# generate data with different ranges
df <- data.frame(
a = sample(seq(0, 2, length.out=20)),
b = sample(seq(100, 500, length.out=20)))
# view data
glimpse(df)## Rows: 20
## Columns: 2
## $ a <dbl> 1.3684211, 1.1578947, 1.2631579, 2.0000000, 0.7368421, 1.5789474, 0.0000000, 1.05…
## $ b <dbl> 331.5789, 457.8947, 394.7368, 184.2105, 247.3684, 310.5263, 121.0526, 100.0000, 1…# data ranges to see if normalization is needed
sapply(df, range)## a b
## [1,] 0 100
## [2,] 2 500# ranges are different => normalize
# Apply min-max and standardization
nrm <- df %>%
mutate(a_MinMax = (a - min(a)) / (max(a) - min(a)),
b_MinMax = (b - min(b)) / (max(b) - min(b)),
a_ZScore = (a - mean(a)) / sd(a),
b_ZScore = (b - mean(b)) / sd(b)
)
# ranges for normalized data
sapply(nrm, range)## a b a_MinMax b_MinMax a_ZScore b_ZScore
## [1,] 0 100 0 0 -1.605793 -1.605793
## [2,] 2 500 1 1 1.605793 1.605793# plots
par(mfrow=c(1,3))
boxplot(nrm[, 1:2], main = 'Original')
boxplot(nrm[, 3:4], main = 'Min-Max')
boxplot(nrm[, 5:6], main = 'Z-Score')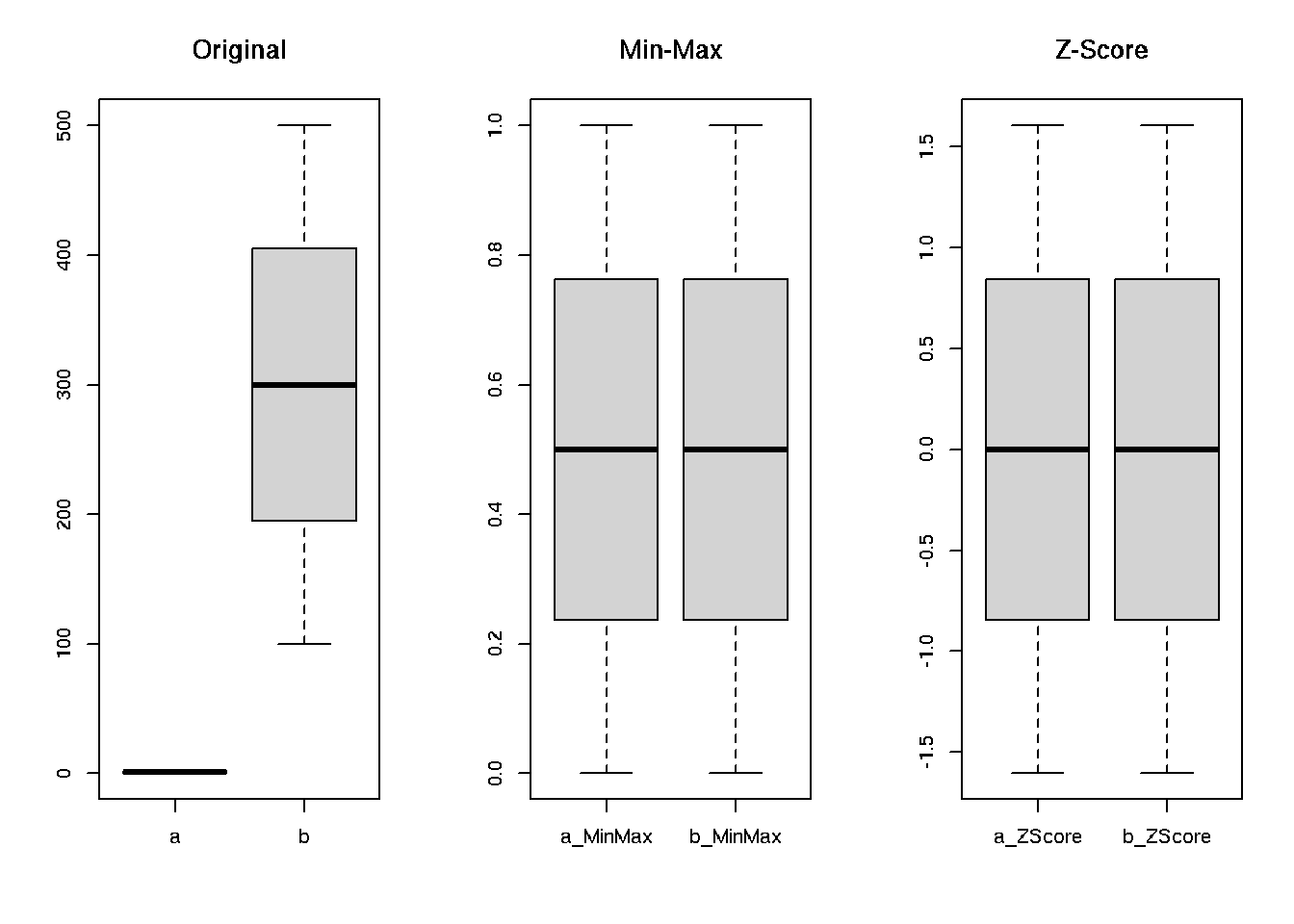
8.1 Normality test
It is possible to use histogram to estimate normality of the distribution.
# QQ-plot - fit normal distibution
qqnorm(v); qqline(v)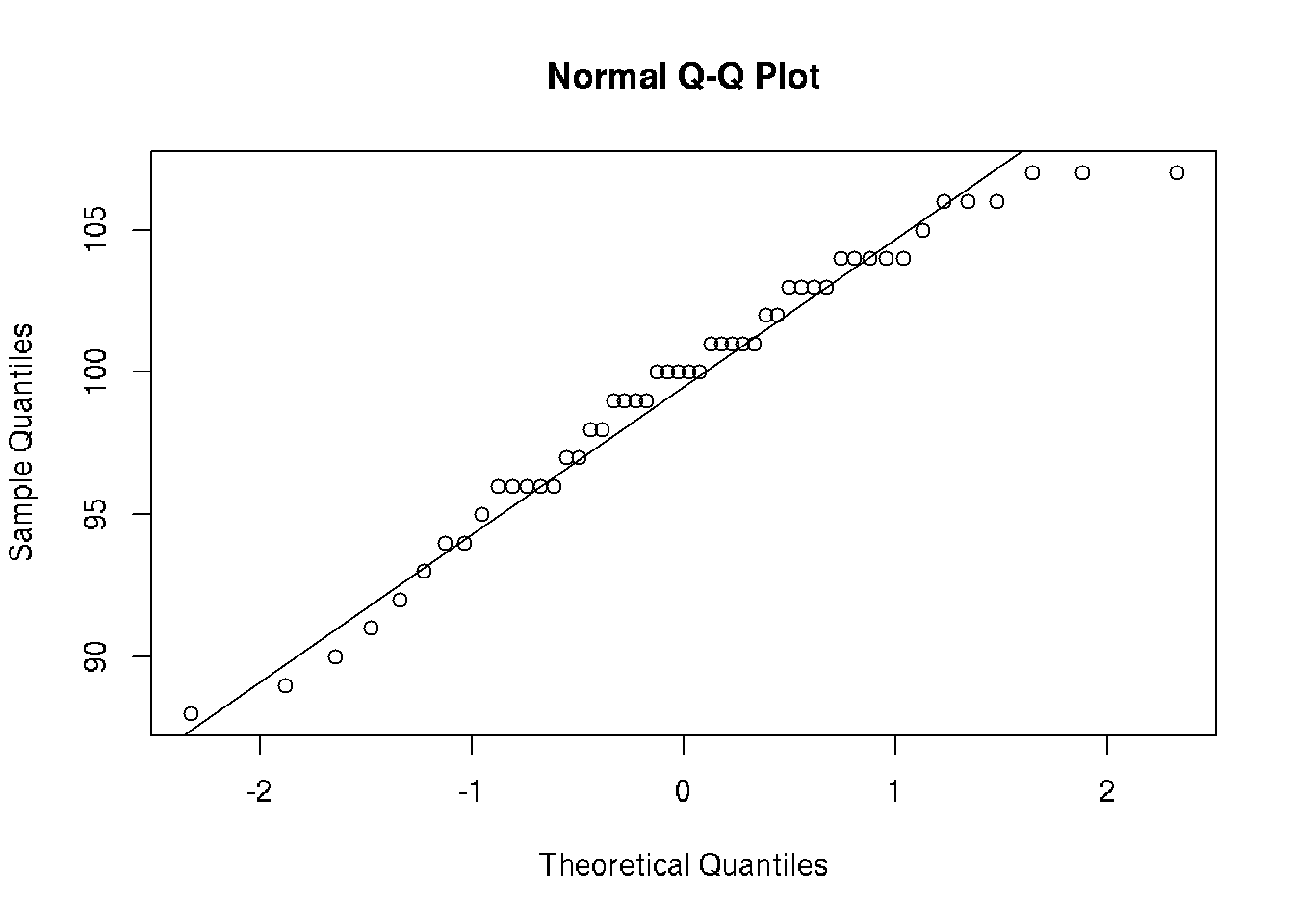
var(v) # variance: sd = sqrt(var)## [1] 23.76367sd(v) # standard deviation## [1] 4.8748sd(v)/sqrt(length(v)) # standard error sd/sqrt(n)## [1] 0.6894008# Z-score (standartization)
# transform distribution to mean=0, variance=1
# z = (x - mean(n))/sd
vs <- scale(v)[,1]
vs## [1] -1.75186680 0.29949948 0.50463610 -2.16214006 0.91490936 -0.11077378 -0.72618366
## [8] 0.70977273 1.53031924 0.09436285 0.50463610 0.29949948 -0.93132029 0.29949948
## [15] -0.72618366 1.12004599 -0.11077378 -0.72618366 0.29949948 -1.13645692 -0.72618366
## [22] -0.11077378 -1.13645692 -1.34159355 0.70977273 0.09436285 0.09436285 0.29949948
## [29] 1.53031924 -0.31591041 1.32518262 -0.52104703 -1.95700343 1.32518262 0.70977273
## [36] -0.11077378 0.91490936 0.91490936 0.91490936 1.32518262 0.70977273 0.09436285
## [43] -2.36727668 1.53031924 -0.31591041 -0.72618366 -1.54673017 0.91490936 0.09436285
## [50] -0.52104703par(mfrow=c(1,2))
hist(v, breaks=10)
hist(vs, breaks=10)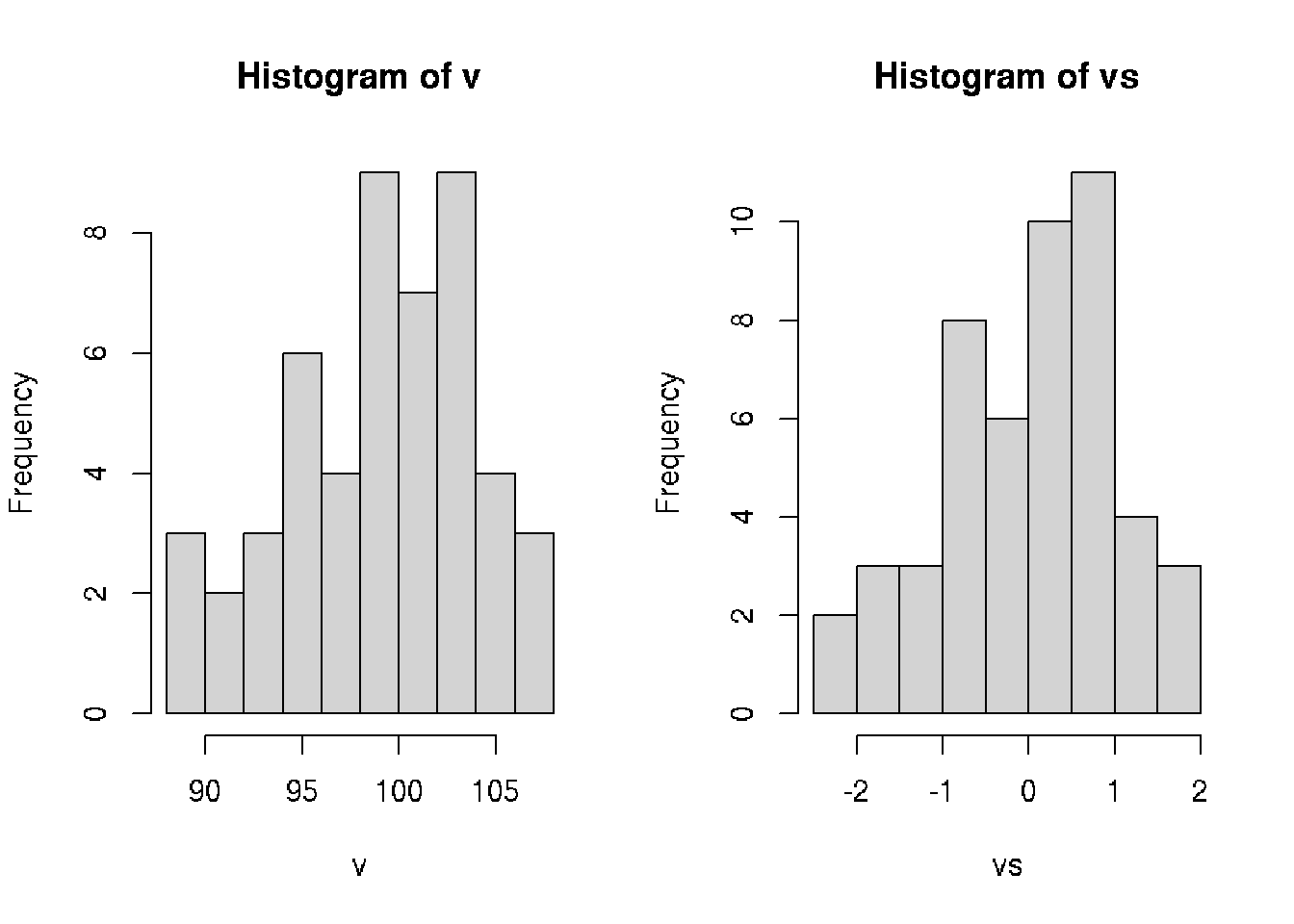
8.2 Finding Confidence intervals
# sample of random integers
x <- round(rnorm(n=50, sd=5, mean=100))
# Confidence interval for normal distribution with p=0.95
m <- mean(x)
s <- sd(x)
n <- length(x)
error <- qnorm(0.95)*s/sqrt(n)
confidence <- c(m-error, m+error)
confidence## [1] 99.16802 101.19198# Confidence interval for t-distribution with p=0.95
a <- 5
s <- 2
n <- 20
error <- qt(0.975,df=n-1)*s/sqrt(n)
# confidence interval
c(left=a-error, right=a+error)## left right
## 4.063971 5.936029Sources
Practicing Machine Learning Interview Questions in R on DataCamp